Overview
Our research focuses on gene regulation in immune cells. Fighting infection requires that diverse cells respond appropriately to pathogens and immune signaling molecules. Immune-related diseases often involve inappropriate responses to these same stimuli. At the level of gene transcription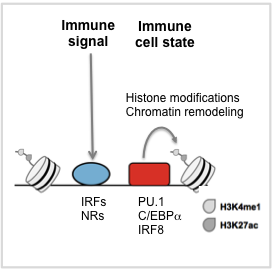 , responses involve coordinated action of transcription factors (TFs) that transmit signaling events (i.e., signal-activated TFs) or that define cell identity and cell polarization (i.e., cell-state TFs) (see Figure). Our approach to a systems-level understanding of immune cell responses is through integrative biophysical and genomic study of signal-activated and cell-state TFs. Our work combines high-throughput biophysical and functional studies with genomics datasets to examine how TFs coordinate gene regulation in response to immunological signals. It is our vision that biophysical understanding of TF function combined with genome-scale data will provide generalizable models needed to understand immune signaling in healthy and disease states.
, responses involve coordinated action of transcription factors (TFs) that transmit signaling events (i.e., signal-activated TFs) or that define cell identity and cell polarization (i.e., cell-state TFs) (see Figure). Our approach to a systems-level understanding of immune cell responses is through integrative biophysical and genomic study of signal-activated and cell-state TFs. Our work combines high-throughput biophysical and functional studies with genomics datasets to examine how TFs coordinate gene regulation in response to immunological signals. It is our vision that biophysical understanding of TF function combined with genome-scale data will provide generalizable models needed to understand immune signaling in healthy and disease states.
Current Projects
A. Specificity of signal-activated TFs
We are currently studying TF specificity of the interferon regulatory factors (IRFs) and the type II Nuclear Receptors (NRs) that mediate anti-viral and metabolic signaling in monocytes, respectively. We are taking a systems approach to examine how members of these TF families discriminate themselves from each other.
Interferon regulatory factor 3 (IRF3), IRF5 and IRF7
The IRFs are critical to the pathogen-recognition response in monocytes and macrophages. Despite similar reported DNA-binding b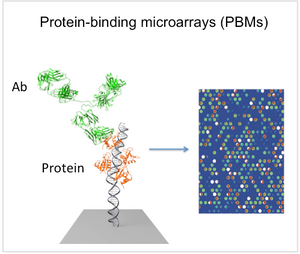 ehavior, these IRFs regulate overlapping yet distinct target gene sets. We recently published a study that used protein-binding microarrays (PBMs) and functional reporter studies to examine the DNA binding and functional specificity of IRF3 IRF5 and IRF7 (Andrilenas et al. (2018) NAR). PBMs (see Figure) allow for the high-throughput characterization of protein-DNA binding specificity. We identified key differences in the IRF-DNA binding and related these binding differences to factor-specific regulation of the type I IFN genes. Unexpectedly, we found an absence of IRF5 binding to all IFN VREs from human and mouse, demonstrating evolutionary pressure specifically against the IRF5 homodimer binding, and suggesting a role for heterodimerization with other family members in type I IFN regulation (see Figure). We also found that the DNA
ehavior, these IRFs regulate overlapping yet distinct target gene sets. We recently published a study that used protein-binding microarrays (PBMs) and functional reporter studies to examine the DNA binding and functional specificity of IRF3 IRF5 and IRF7 (Andrilenas et al. (2018) NAR). PBMs (see Figure) allow for the high-throughput characterization of protein-DNA binding specificity. We identified key differences in the IRF-DNA binding and related these binding differences to factor-specific regulation of the type I IFN genes. Unexpectedly, we found an absence of IRF5 binding to all IFN VREs from human and mouse, demonstrating evolutionary pressure specifically against the IRF5 homodimer binding, and suggesting a role for heterodimerization with other family members in type I IFN regulation (see Figure). We also found that the DNA 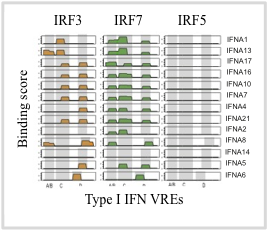 sequence of binding sites can modulate activity independently of binding affinity, suggesting that DNA-based allostery is critical to IRF function. Allostery remains a poorly characterized phenomenon for TFs, and in this work we demonstrate clearly for the IRFs how changes in specific positions of the binding site can enhance activity at the expense of binding affinity. We are currently using massively parallel reporter assays (MPRAs), in conjunction with PBMs, to examine this regulatory logic of IRF dimers.
sequence of binding sites can modulate activity independently of binding affinity, suggesting that DNA-based allostery is critical to IRF function. Allostery remains a poorly characterized phenomenon for TFs, and in this work we demonstrate clearly for the IRFs how changes in specific positions of the binding site can enhance activity at the expense of binding affinity. We are currently using massively parallel reporter assays (MPRAs), in conjunction with PBMs, to examine this regulatory logic of IRF dimers.
Type II Nuclear Receptors (NRs)
Type II NRs are signal-activated TFs that share a common partner – the Retinoid X Receptor (RXR) – and play key roles in immunity and metabolism. NRs bind diverse endogenous ligands and many therapeutic drugs. Despite responding to distinct ligands, overlap in NR target genes and genome-wide binding reveals significant crosstalk between NR signaling pathways. Clarifying mechanisms of NR crosstalk and specificity is critical to understanding the effects of both endogenous ligands and therapeutic drugs. We are currently finishing a comprehensive PBM-based study of the DNA-binding for twelve NR heterodimers. Our study has revealed considerable overlap in the NR DNA binding while also highlighting key differences between dimers – challenging our current view of NR binding specificity.
B. Specifying cell state in human monocytes
Lineage factors are TFs that define cell identity by establishing the cell type-specific enhancer landscape. Many immune cells also adopt polarization states, such as pro-inflammatory M1 macrophages versus anti-inflammatory M2 macrophages. Polarization states are similarly defined by TFs that modify gene expression and the enhancer landscape (e.g., Foxp3 in regulatory T cells). We aggregately refer to these TFs as ‘cell-state TFs’. Understanding how cell-state TFs integrate with signal-activated TFs is central to understanding cell type-specific response of immune cells. We are currently studying factors that affect the binding and function of the monocyte/macrophage lineage factors PU.1, C/EBPa and IRF8. We are presently examining role for DNA-binding affinity and cooperative protein complexes in defining the PU.1 cistrome in human monocytes
C. Technology development
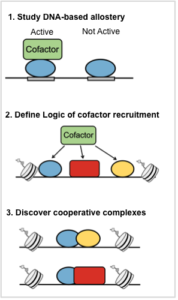
We continue to develop the PBM technology for the biophysical characterization of TFs. We are working to develop PBM-based approaches to (see Figure) : (1) Study allostery, (2) Define the logic of cofactor recruitment at enhancers, (3) discover and characterize cooperatively binding TFs.